Tan Lab
- Research
- Members
- Publications
- Contact Us
Our lab combines cutting-edge omics tools with 'wet' lab approaches to study transcriptional and post-transcriptional regulation in normal development and its disruption in disease, with the goal of advancing treatments for human infertility and neurodevelopmental disorders.
Our research interests overlap with the following fields:
SSC generation and development
Spermatogonial stem cells (SSCs) are the adult stem cells of the testis that sustain spermatogenesis and, ultimately, male fertility. Like other stem cells, SSCs balance self‑renewal with differentiation. Upon differentiation, they give rise to successive spermatogenic cell types that eventually produce sperm. SSCs are of broad interest both as a model for fundamental stem cell biology and as a potential therapeutic platform for treating male infertility—particularly for men with non‑obstructive azoospermia and pediatric cancer survivors exposed to gonadotoxic therapies.
Our laboratory investigates the cellular and molecular mechanisms that govern human and mouse SSC formation and development. Using single‑cell RNA sequencing (scRNA‑seq), bulk RNA-seq, and marker‑based approaches, we have been uncovering the complexity of spermatogonia—their heterogeneity, lineage relationships, and molecular logic (Cell Rep 2019; Development 2020; PNAS 2020; Cell Rep 2026, in press).
Another major focus of the lab is the homeobox transcription factor RHOX10, a critical regulator of SSC genesis and development. We are not only defining the molecular mechanisms by which RHOX10 drives SSC formation and development (Cell Rep 2021; manuscript in preparation), but also uncovering its role in safeguarding genome integrity (PNAS 2021).
Our current efforts aim to build transcriptional regulatory networks that orchestrate SSC formation and developmental progression. To achieve this, we integrate a broad suite of omics and functional tools—including RNA‑seq, scRNA‑seq, CUT&Tag, SLAM‑seq, CRISPR perturbations, “mimic and rescue” strategies, and mouse genetics. Through these approaches, we seek to uncover the fundamental principles that define SSC identity, function, and therapeutic potential.
Nonsense-mediated RNA decay (NMD)
RNA turnover regulation, a crucial counterpart to transcriptional regulation, plays an essential role in maintaining cellular homeostasis and facilitating dynamic responses to environmental and developmental cues. Despite its importance, studies on RNA turnover regulation are only beginning to grow compared to the extensive research on transcriptional regulation. I have pioneered studies examining how RNA turnover and transcription jointly shape gene expression during mouse germline specification, identifying distinct regulatory categories that reveal how RNA turnover and transcription interplay to dictate steady‑state RNA levels. I also demonstrated that RNA turnover regulation alone plays a critical role in driving germline specification (Nucleic Acids Res 2022).
Beyond this broad regulatory question, our lab has focused on the nonsense‑mediated RNA decay (NMD) pathway to elucidate its physiological roles in development. My first project in this area examined NMD function in the nervous system using Upf3b‑null mice (eLife 2020). Through single‑cell RNA‑seq, bulk RNA‑seq, and RiboTag profiling of olfactory sensory neurons, we demonstrated that UPF3B is essential for multiple aspects of olfactory neural development—including neuronal maturation and establishment of the olfactory receptor repertoire—providing early evidence that NMD contributes directly to neural circuit formation.
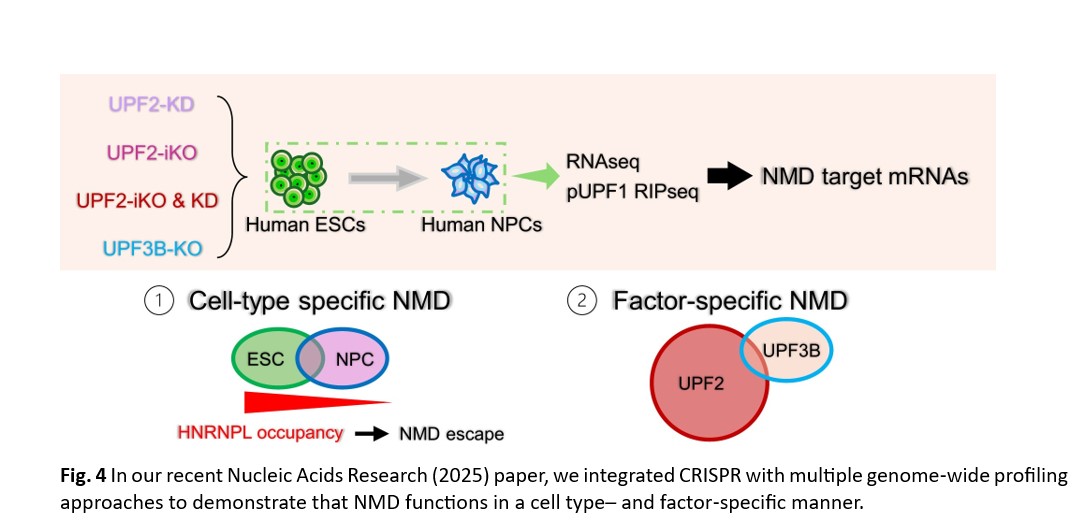
Building on this foundation, I conceived and conducted a study (Nucleic Acids Res 2025) generating conditional UPF2‑depleted and UPF3B‑knockout human embryonic stem cells (hESCs) and their differentiated neural progenitor cells (hNPCs). Using RNA‑seq and phospho‑UPF1 RIP‑seq, I identified high‑confidence NMD targets in both cell types. These analyses revealed that NMD is not a rigid pathway degrading a fixed set of mRNAs, but rather a flexible mechanism whose targets depend on cellular context and the presence or absence of specific protein factors. I further discovered that NMD selects mRNAs for degradation in a factor‑specific manner, revising long‑standing models of NMD substrate recognition and providing a framework for understanding how NMD dysfunction contributes to neurodevelopmental disorders.
In a separate project (Science Adv 2025), I studied a 442‑nt noncoding element within SMG6, one of the key NMD factors. This element, HAR123, is deeply conserved across mammals and marsupials yet has diverged rapidly in the human lineage. I demonstrated that HAR123 functions as a neural enhancer that promotes human hNPC formation. Comparative analyses revealed that the human and chimpanzee orthologs differentially regulate neural differentiation programs. I identified direct HAR123 targets and showed that HIC1 acts downstream of HAR123 to promote NPC formation. HAR123‑knockout mice exhibited a specific defect in cognitive flexibility and an altered neural–glia ratio in select hippocampal regions—findings with implications for neurodevelopmental disorders, many of which involve disrupted neural–glia balance.
Our current research focuses on: (i) studying the physiological roles of regulated RNA turnover in normal development, and (ii) defining how NMD factors shape stem cell fate and function across specific developmental contexts, and how their dysregulation contributes to human disorders.