Wilkinson Lab
- About
- Research
- Lab Members
- Publications
- Contact Us
In particular, our laboratory is interested in how transcriptional and post-transcriptional pathways control normal development and how their disruption causes disease.
Our work is laying the groundwork for curing specific cases of infertility, cystic fibrosis, and neurodevelopmental disorders.
Our research interests overlap with the following fields:
o Stem cell biology
o Reproductive biology
o Developmental biology
o Neurobiology
o RNA biology
o Transcriptional regulation
Infertility is an extremely common human condition. One in seven couples fail to conceive after 1 yr of unprotected sex. A large set of candidate genes involved in infertility are those in a gene cluster that we discovered on the X chromosome (MacLean et al. Cell 2005). These Rhox genes encode transcription factors expressed selectively in the male and female reproductive tract. Our recent focus has been on one particular member of the Rhox gene cluster—Rhox10—which we discovered drives the initial formation of spermatogonial stem cells (SSCs) (Song et al. Cell Rep 2016). There is considerable interest in SSCs, as they are the stem cells in the testis that are essential for spermatogenesis. Understanding how SSCs form in vivo can lead to methods to generate and expand SSCs in vitro for treating male infertility. Towards this goal, we have identified RHOX10 gene targets using a variety of “omics” approaches (CUT&Tag, RNAseq, and SLAMseq), and defined a molecular circuit involving RHOX10 and two downstream transcription factors that promotes SSC formation (Tan et al. Cell Reports 2021) (Fig. 1). We also discovered that RHOX10 has a second function – it defends the male germline from mutations engendered by transposable elements (Tan et al. PNAS 2021). Our future studies are focused on defining gene networks essential for SSC establishment and transposon defense.
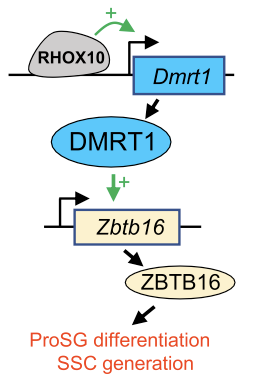
Fig. 1. A transcription factor circuit driving germ cell differentiation (Tan et al. Cell Reports 2021). ProSG, Pro-Spermatogonia; SSC, spermatogonial stem cell
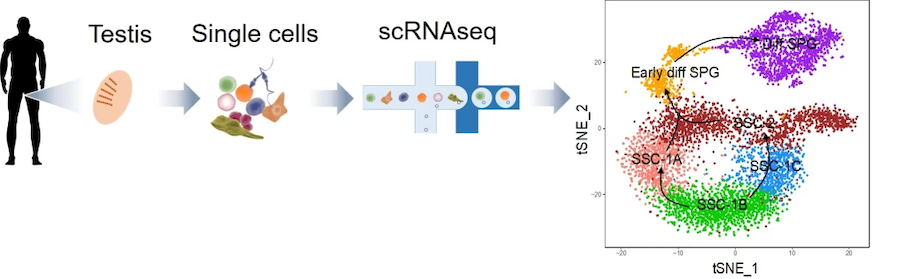
Fig. 2. Single-cell RNA-sequencing (scRNAseq) analysis defines human spermatogonial cell subsets (Sohni et al. Cell Reports 2019).
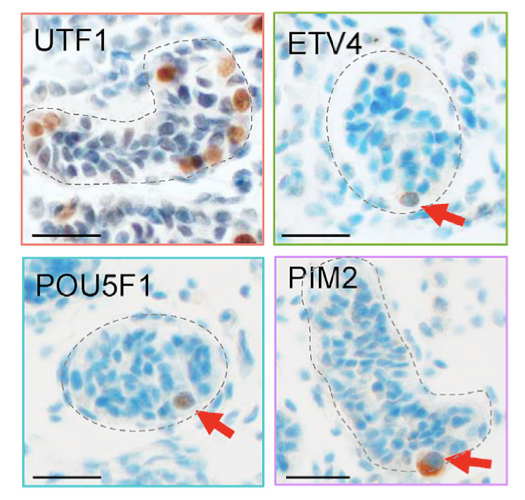
Fig. 3. Protein markers identified by scRNAseq analysis that mark primordial germ cell (PGC)-like cells in newborn human testes (Sohni et al. Cell Reports 2019). ETV4, PIM2, and POU5F1 are PGCL markers we defined, whereas UTF1 is a previously-defined broader immature germ-cell marker.
Towards the ultimate goal of developing “SSC therapy approaches” to treat male infertility, we have used scRNAseq analysis to identify undifferentiated spermatogonia subsets that have the molecular characteristic of SSCs (Sohni et al. Cell Rep 2019, Tan et al. Dev 2020). This goal has culminated most recently in the definitive identification of highly enriched human SSC population (defined using xenograft germ-cell transplantation) bearing a specific cell-surface protein (Tan et al. PNAS 2020). We have use these purified human SSCs to define both the “human SSC transcriptome” and approaches to both differentiate and culture undifferentiated spermatogonia with the characteristics of human SSCs (Tan et al. PNAS 2020).
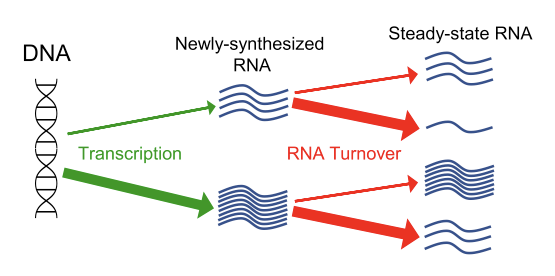
Fig. 4. The steady-state level of a given mRNA is dictated by both its rate of synthesis (transcription) and its decay rate. See our recent analysis of these two processes genome-wide (Tan et al. Nucleic Acids Res 2022).
Thus, the current focus on transcriptional pathways controlling developmental processes is incomplete. We are interested in the developmental roles of the regulated RNA turnover pathway NMD. This highly conserved and selective RNA decay pathway has captured the imagination of both basic scientists and clinicians. NMD degrades specific RNAs in its capacity as a quality control mechanism and as a means to control diverse biological processes. For more than 20 years, we have studied NMD, including its regulation, its evolution, and its roles in different biological processes. One process involving NMD is early embryonic development, which has led to clinical interest in NMD’s possible role in aberrant pregnancies, including miscarriages. Towards understanding NMD’s role in early embryonic development, we have reported: (i) functions for NMD in human embryonic stem cell development (Lou et al. Stem Cell Reports 2016), (ii) shown that that loss of the NMD factor UPF3A causes embryonic lethality in mice (Shum et al. Cell 2016), and (iii) done in-depth phenotypic and molecular analyses of NMD-deficient mice embryos. NMD also functions in neural development, neuron function, and cognition (Jaffrey & Wilkinson Nature Reviews Neuroscience 2018). Several groups have demonstrated that one particular NMD gene—UPF3B—when mutated, causes intellectual disability (and is associated with schizophrenia and autism) in humans, leading to efforts to manipulate NMD to treat neurological diseases. Towards this end, we generated a mouse model harboring a Upf3b-null mutation and showed these NMD-deficient mice have specific behavioral and neural defects (Huang et al. Mol Psych 2018). We also identified roles for NMD in neural differentiation (Bruno et al. Mol Cell 2011; Lou et al. Cell Rep 2014; Huang et al. Mol Psych 2018) and showed that NMD has specific functions in olfactory sensory neurons (Tan et al. eLIFE 2020). We have also asked broader questions about RNA turnover regulation, including its relative role as compared with transcriptional regulation (Tan et al. Nucleic Acids Res 2022). Our current studies are focused on (i) defining roles for the NMD factor paralogs—UPF3A and UPF3B—in specific developmental contexts, (ii) manipulating NMD to treat genetic diseases (see below), and (iii) determining whether NMD has roles in human evolution (see below).
This devastating genetic disease leads to severe damage of several organs, with fatality typically occurring because of lung damage. The gene responsible for cystic fibrosis—CFTR—was identified over 30 years ago, and drugs have been recently been approved that act on mutant CFTR proteins to provide significant therapeutic benefit to many cystic fibrosis patients. However, cystic fibrosis patients harboring nonsense (stop codon) mutations in the CFTR gene are almost universally unresponsive to these drugs. Thus, we have recently obtained grant support to develop strategies to treat these non-responsive patients. One approach we are focused on is to suppress the NMD pathway as a means to stabilize the nonsense-mutant CFTR mRNA. By combining this NMD suppression therapy with nonsense codon-readthrough drugs, we have had success in dramatically upregulating full-length CFTR protein in bronchial cells harboring nonsense mutations in the CFTR gene.
A fundamental question in biology is: how did humans acquire their unique characteristics? What allows us to stand upright, while our primate ancestors walked on all fours? What brain alterations drove our increased intelligence and allowed us to perceive our own mortality? In our laboratory, we are investigating whether some human-specific traits are driven by alterations in NMD during primate evolution. We were led to this possibility because we identified conserved sequences in and around NMD genes that are rapidly evolving in humans. These human accelerated regions (HARs) are segments of DNA that have two specific qualities that—together—make them prime candidates for specifying human-specific traits (https://www.the-scientist.com/features/decoding-human-accelerated-regions-33114). First, they are highly conserved throughout vertebrates. This quality means that HARs are likely to specify functions that have been maintained for ~500 million years. Second, they are evolving much more rapidly in humans than other species. This second characteristic means HARs are candidates to be responsible for driving evolutionary alterations unique to humans. We have identified one particular HAR in a NMD gene that our studies indicate is a transcriptional enhancer critical for neural development. Our future goals include determining whether this HAR regulates NMD and whether its evolution conferred unique neural properties to humans. This project provides an exciting opportunity to address a fundamental question in biology that currently remains largely a black box.